Ch. 6 Predictive Modeling
6.1 Overview
This example uses data from a reaction time experiment where observers searched for a green circle amongst rectangles, and were asked to report if a line inside the green circle was vertical or horizontal. On some trials, a red rectangle was present, i.e, a distractor. The experiment was conducted in two ways: (1) a random ordering of distractor present and absent trials, and (2) a blocked ordering of distractor present and absent trials. With blocked administration, presumably participants had time to habituate to the distractor and would have a smaller response time decrement as compared with the random presentation condition.
6.3 Load data
exp_df <- read_csv("data/exp1.csv")## Rows: 56896 Columns: 11
## ── Column specification ─────────────────────────────────────────────────────────────────
## Delimiter: ","
## chr (6): practice, datetime, distractor, isRandom, PARTICIPANT, BLOCK
## dbl (5): count_trial_sequence, correct, display_size, response_time, RT
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# show data structure
head(exp_df)## # A tibble: 6 × 11
## practice count_trial_sequence datetime correct display_size distractor
## <chr> <dbl> <chr> <dbl> <dbl> <chr>
## 1 yes 0 11/30/17 15:38 0 5 present
## 2 yes 1 11/30/17 15:38 0 5 present
## 3 yes 2 11/30/17 15:38 0 5 present
## 4 yes 3 11/30/17 15:38 0 5 present
## 5 yes 4 11/30/17 15:38 1 5 present
## 6 yes 5 11/30/17 15:38 0 5 present
## # … with 5 more variables: response_time <dbl>, isRandom <chr>,
## # PARTICIPANT <chr>, RT <dbl>, BLOCK <chr>6.4 Pre-process data
6.4.1 Create additional columns, remove columns
exp_pp_ = exp_pp %>%
mutate(is_practice = ifelse(practice == "yes", 1, 0)) %>%
mutate(TRIAL = count_trial_sequence) %>%
select(-count_trial_sequence, -response_time, -practice) %>%
select(datetime, PARTICIPANT, BLOCK, TRIAL, is_practice, correct, RT, everything()) %>%
mutate(EXP_BLOCK_RANDOM = ifelse(isRandom == "blocked", 0, 1))6.5 Produce simple aggregates
# overall means and record count by participant
part_means = exp_pp_ %>%
group_by(PARTICIPANT, datetime) %>%
summarise(mean_rt = mean(RT, na.rm=T),
sd_rt = sd(RT, na.rm=T),
q10_rt = quantile(RT, probs=0.1, na.rm=T),
q90_rt = quantile(RT, probs=0.9, na.rm=T),
n_correct = sum(correct),
n = n()) %>%
inner_join(metadata)## `summarise()` has grouped output by 'PARTICIPANT'. You can override using the `.groups`
## argument.
## Joining, by = c("PARTICIPANT", "datetime")
# overall means by block
part_means_byblock = exp_pp_ %>%
group_by(PARTICIPANT, datetime, BLOCK, is_practice) %>%
summarise(mean_rt = mean(RT, na.rm=T),
sd_rt = sd(RT, na.rm=T),
q10_rt = quantile(RT, probs=0.1, na.rm=T),
q90_rt = quantile(RT, probs=0.9, na.rm=T),
n_correct = sum(correct),
n = n()) %>%
inner_join(metadata)## `summarise()` has grouped output by 'PARTICIPANT', 'datetime', 'BLOCK'. You can override
## using the `.groups` argument.
## Joining, by = c("PARTICIPANT", "datetime")6.5.1 Mean center the reaction times
exp_m = exp_pp_ %>%
inner_join(part_means) %>%
mutate(imean_c_RT = RT - mean_rt)## Joining, by = c("datetime", "PARTICIPANT", "EXP_BLOCK_RANDOM")6.6 Data visualization
ggplot(part_means_byblock %>% filter(!is_practice), aes(BLOCK, mean_rt, group=BLOCK, color=EXP_BLOCK_RANDOM)) +
geom_boxplot() +
facet_grid(. ~ EXP_BLOCK_RANDOM) +
theme_bw() +
labs(x= "Experiment Block", y = "Mean Response Time (ms)")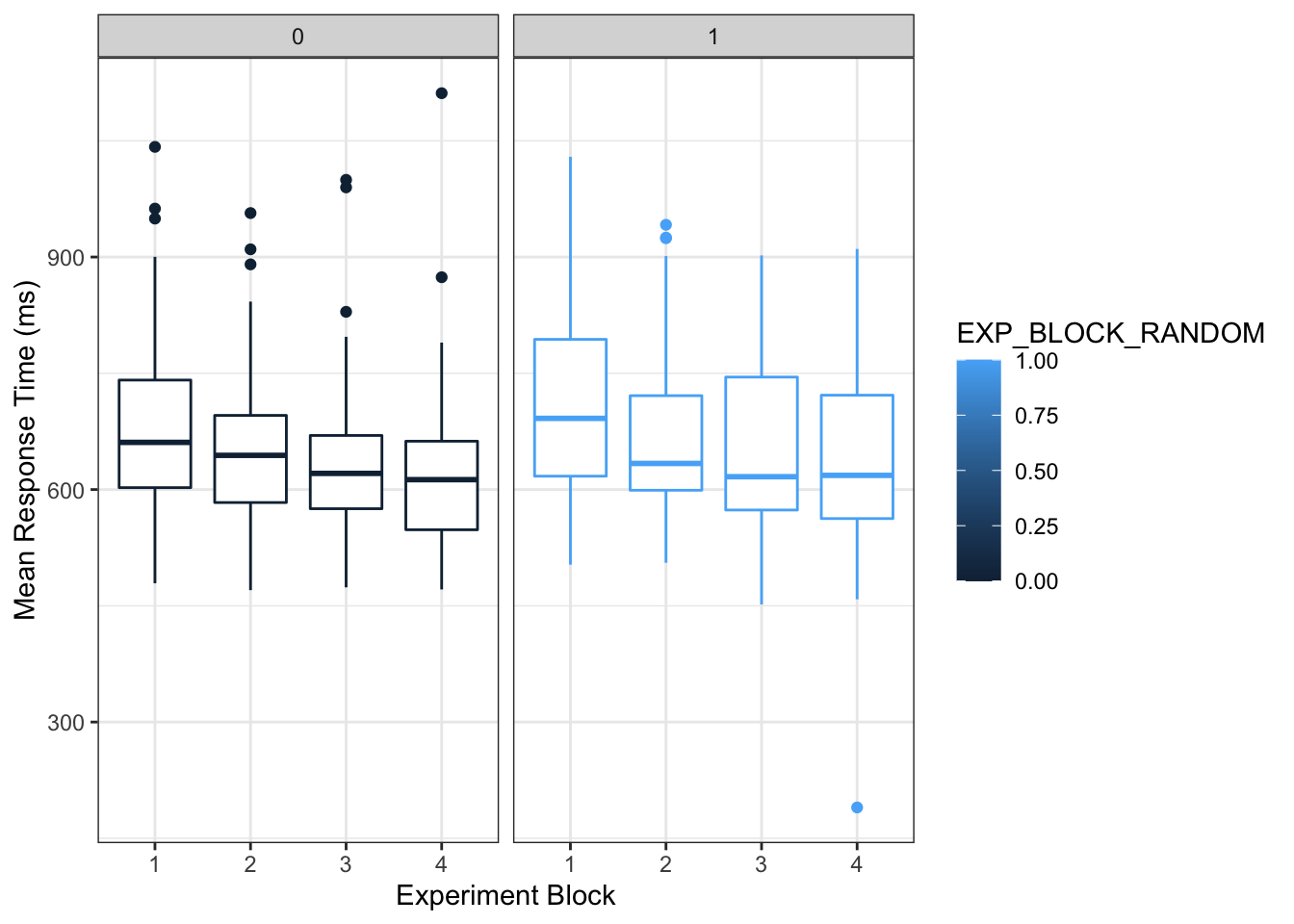
6.8 Run simple regression on block means
##
## Call:
## lm(formula = mean_rt ~ n_correct, data = part_means_byblock)
##
## Residuals:
## Min 1Q Median 3Q Max
## -626.20 -118.35 -30.63 84.85 964.95
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 811.3833 9.5612 84.86 <2e-16 ***
## n_correct -1.6608 0.1424 -11.67 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 191.6 on 1014 degrees of freedom
## Multiple R-squared: 0.1183, Adjusted R-squared: 0.1175
## F-statistic: 136.1 on 1 and 1014 DF, p-value: < 2.2e-166.8.1 Show resulting model equation
# show model equation
library(equatiomatic)
extract_eq(fit, use_coefs=TRUE, wrap=TRUE)\[ \begin{aligned} \operatorname{\widehat{mean\_rt}} &= 811.38 - 1.66(\operatorname{n\_correct}) \end{aligned} \]
6.9 Run as series of incremental multilevel model
# run models, starting with unconditional means model
fitm_0 = lme4::lmer(RT ~ 1 + (1|PARTICIPANT),
data=exp_pp_)
fitm_1 = lme4::lmer(RT ~ TRIAL + (1|PARTICIPANT),
data=exp_pp_)
fitm_2 = lme4::lmer(RT ~ TRIAL + is_practice + (1|PARTICIPANT),
data=exp_pp_)
fitm_3 = lme4::lmer(RT ~ TRIAL + is_practice + correct + (1|PARTICIPANT),
data=exp_pp_)
fitm_4 = lme4::lmer(RT ~ TRIAL + is_practice + correct +
distractor +
(1|PARTICIPANT),
data=exp_pp_)
fitm_5 = lme4::lmer(RT ~ TRIAL + is_practice + correct +
distractor +
display_size +
EXP_BLOCK_RANDOM +
(1|PARTICIPANT),
data=exp_pp_)6.9.1 Compare models
# compare models
anova(fitm_0, fitm_1, fitm_2, fitm_3, fitm_4, fitm_5)## refitting model(s) with ML (instead of REML)## Data: exp_pp_
## Models:
## fitm_0: RT ~ 1 + (1 | PARTICIPANT)
## fitm_1: RT ~ TRIAL + (1 | PARTICIPANT)
## fitm_2: RT ~ TRIAL + is_practice + (1 | PARTICIPANT)
## fitm_3: RT ~ TRIAL + is_practice + correct + (1 | PARTICIPANT)
## fitm_4: RT ~ TRIAL + is_practice + correct + distractor + (1 | PARTICIPANT)
## fitm_5: RT ~ TRIAL + is_practice + correct + distractor + display_size + EXP_BLOCK_RANDOM + (1 | PARTICIPANT)
## npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
## fitm_0 3 775163 775190 -387579 775157
## fitm_1 4 772908 772944 -386450 772900 2256.615 1 < 2.2e-16 ***
## fitm_2 5 771229 771274 -385610 771219 1681.167 1 < 2.2e-16 ***
## fitm_3 6 770470 770523 -385229 770458 761.500 1 < 2.2e-16 ***
## fitm_4 7 770331 770394 -385159 770317 140.332 1 < 2.2e-16 ***
## fitm_5 9 770323 770404 -385152 770305 12.428 2 0.002001 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 16.10 Create publication-ready table of results
sjPlot::tab_model(fitm_0, fitm_5)| RT | RT | |||||
|---|---|---|---|---|---|---|
| Predictors | Estimates | CI | p | Estimates | CI | p |
| (Intercept) | 670.44 | 653.24 – 687.63 | <0.001 | 796.63 | 774.02 – 819.25 | <0.001 |
| TRIAL | -0.29 | -0.30 – -0.27 | <0.001 | |||
| is practice | 111.09 | 105.44 – 116.74 | <0.001 | |||
| correct | -98.78 | -105.82 – -91.73 | <0.001 | |||
| distractor [present] | 20.84 | 17.40 – 24.29 | <0.001 | |||
| display size | 1.53 | 0.67 – 2.39 | 0.001 | |||
| EXP BLOCK RANDOM | -6.50 | -28.08 – 15.07 | 0.555 | |||
| Random Effects | ||||||
| σ2 | 47870.23 | 43951.44 | ||||
| τ00 | 9588.24 PARTICIPANT | 9799.13 PARTICIPANT | ||||
| ICC | 0.17 | 0.18 | ||||
| N | 126 PARTICIPANT | 126 PARTICIPANT | ||||
| Observations | 56896 | 56896 | ||||
| Marginal R2 / Conditional R2 | 0.000 / 0.167 | 0.068 / 0.238 | ||||
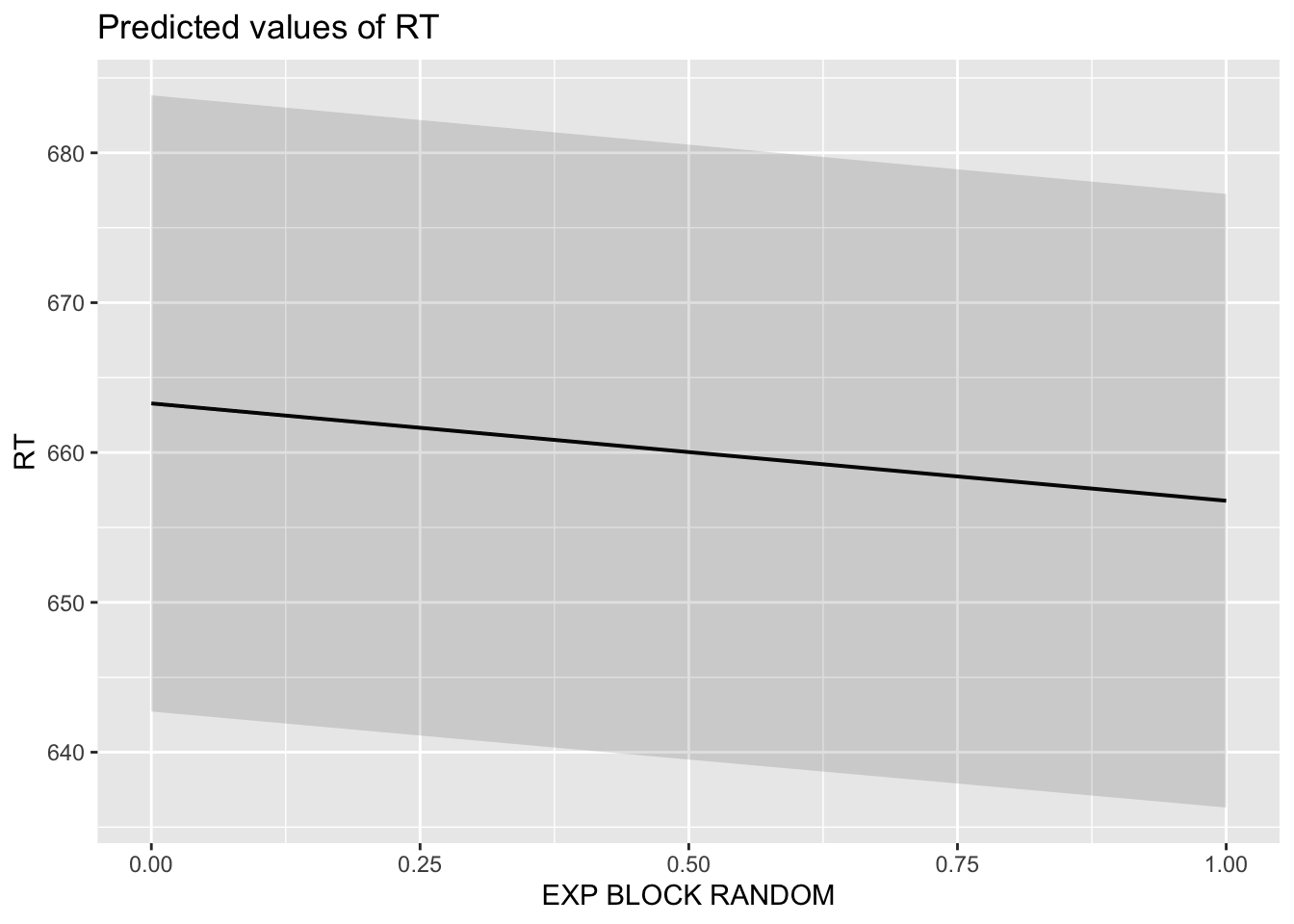
6.11 Create publication-ready figure of results
sjPlot::plot_model(fitm_5, type="pred")## $TRIAL
##
## $is_practice
##
## $correct
##
## $distractor
##
## $display_size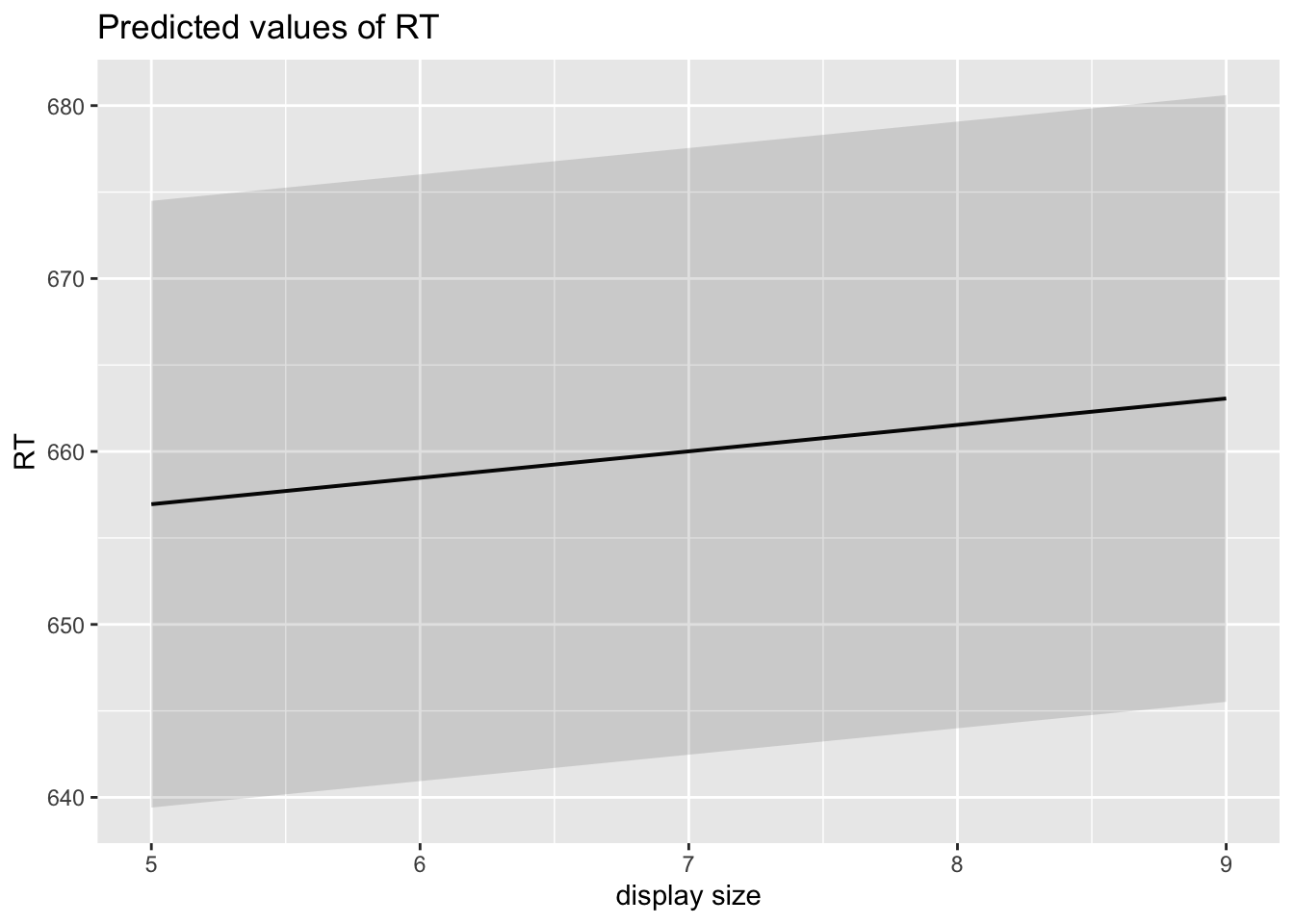
##
## $EXP_BLOCK_RANDOM